I’ve analyzed Ancient Roman mtDNA, again from the NIH Database, and I am unable to find any significant matches in the dataset below, which now consists of 29 global ethnicities, including other ancient ethnicities. Even using BLAST, I am unable to find any matches that don’t contain significant gaps or other changes to the alignment, suggesting that the Ancient Romans were annihilated on the maternal side. Don’t forget, despite the fact that everyone uses Haplogroups to analyze ancestry, it arguably doesn’t make much sense for mtDNA, because it’s passed on from one generation to the next, with basically no mutations at all. As a consequence, you shouldn’t be able to inherit a portion of a genome, and instead, you either inherit the whole thing, or you’re not related at all on the maternal side. We can nonetheless explain the emergence of geographically local mtDNA, by assuming common mutations to an existing line are possible, which would cause people to branch. For example, if population A comes from Asia, and they move to Europe, then the food could be different, and because mtDNA is intimately involved in the production of ATP (i.e., energy), it makes perfect sense that environment would create geographically common mutations. Regardless of this, the bottom line is that I am not aware of any other ethnicity that requires a significant change in alignment to find a close match, including ancient genomes, other than the Neanderthals, who are known to be extinct.
The only meaningful matches between the Ancient Romans and others happen around 30% of matching bases (i.e., requiring 30% of bases to match with the Romans). Anything above that starts to produce an empty set, and so they are otherwise a genetically self-contained group of samples, again, save for significant changes in alignment, which is not necessary for any of the other populations in the dataset. That is, all of the other 28 ethnicities, including several ancient samples, have 99% matches, without any significant changes to the alignment. Even Heidelbergensis finds 97% matches in many modern populations, again, without any changes to alignment. Heidelbergensis is thought to have gone extinct hundreds of thousands of years ago, though they plainly didn’t on the maternal line. Again, this suggests that the Ancient Romans were annihilated, which simply could not have occurred quickly, given the scope of the Roman Empire, and so it must have been deliberate.
As a practical matter, I doubt this many people could have been killed in a short amount of time, and so it doesn’t seem possible that the genocide occurred during the actual Fall of Rome. Instead, I’d wager that it was the result of systematic genocide over centuries, presumably at the hands of the Catholic Church, given that they ruled Continental Europe for centuries, and given their emphasis on backwardness and a lack of progress generally. Insults aside, you can plainly see a decline in the quality of life, art, and thought after the Fall of Rome, suggesting that the population that replaced the Ancient Romans was genetically distinct. In particular, the ability to build large free-standing domes, and the recipe for concrete were both lost.
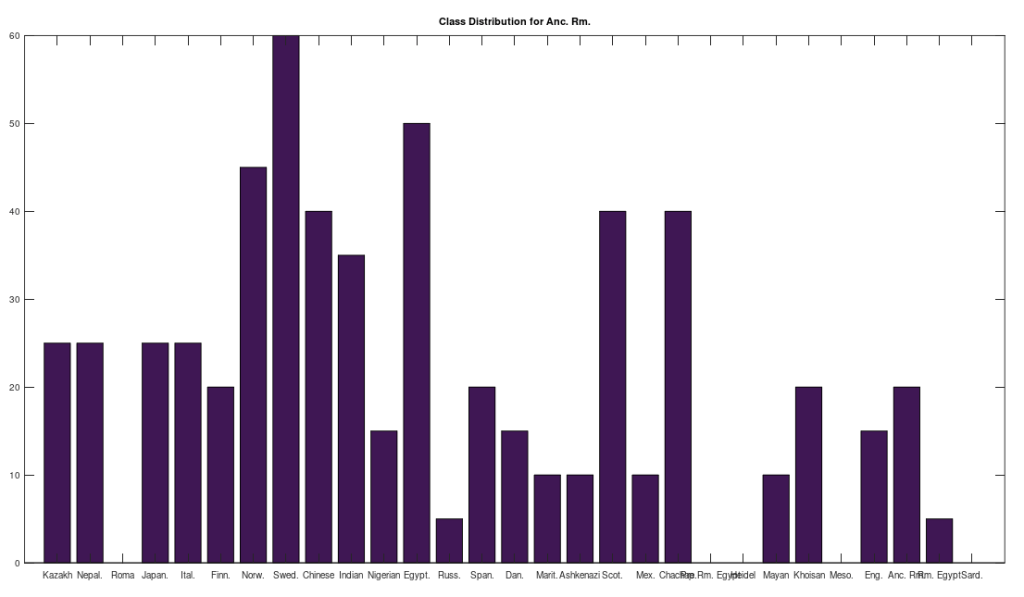
The chart above shows the distribution of matching ethnicities at 30% of matching bases, and you again see a connection to Northern Europe and Asia, as the same is true of the Pre-Roman Egyptians. However, this is a very low threshold, though it is higher than chance, which is 25% of matching bases. This is consistent with the language, since the European languages are generally believed to be derived from early Indian languages. I think the net takeaway is, ancient Europeans seem to come from Asia, and that the Romans were probably annihilated over centuries, possibly by people indigenous to Europe.
Here’s the dataset, which includes links to the NIH Database for every genome:
https://www.dropbox.com/s/ht5g2rqg090himo/mtDNA.zip?dl=0
Discover more from Information Overload
Subscribe to get the latest posts sent to your email.